– Correctly evaluates the stability and reactivity of the shape of molecules, such as of proteins and catalysts –
Researchers) NAKAMURA Takenobu, Senior Researcher, Materials Informatics Team, Research Center for Computational Design of Advanced Functional Materials
- Devised a new formula that overcomes the problems of the conventional formula expressed using only the probability density function
- Enables calculation of free-energy landscape independent of the way molecular shapes are represented
- Contributing to the elucidation of the mechanisms of catalysis and protein folding reactions and to the field of drug discovery
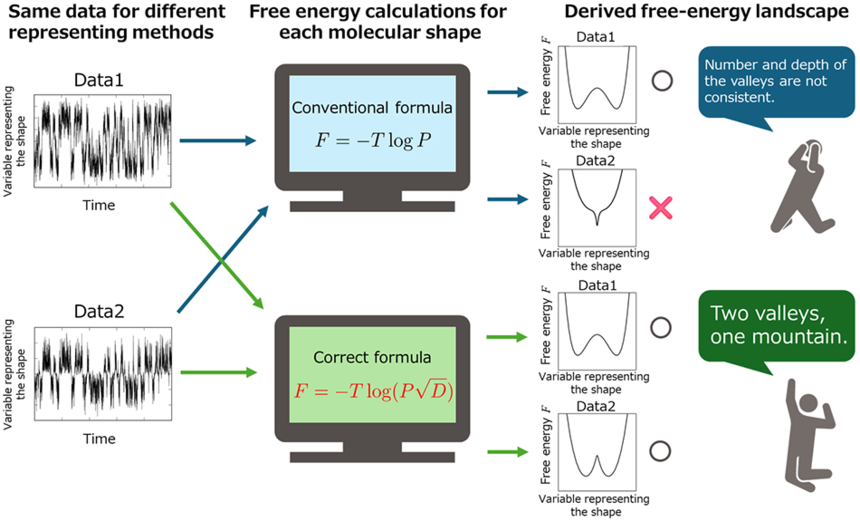
Correct free-energy landscape can evaluate the number and depth of valleys independent of how the molecular shapes are represented.
Free energy is a fundamental physical quantity in describing protein folding and catalysis and indicates the reactivity of a molecule. A protein with a stable shape will have a low free energy. During reactions and folding, the shape of the molecule changes and the free energy fluctuates accordingly. The free-energy landscape is a contour representation of the free energy for each possible shape of the molecule and is an important physical quantity that links the thermodynamic and kinetic properties of a reaction.
However, the conventional definition of free-energy landscape is known to be ill-defined because it does not have the invariance that physical quantities must satisfy. For example, there are several different ways of expressing the same angle, such as the arc length of the unit circle θ [radians] or its cosine χ=cos θ, but the physical property obtained from any of these ways must be invariant. Naturally the free-energy landscape should be invariant regardless of the way the shape of the molecule is represented. However, the conventional definition of free energy gave different results depending on which display method was used such as whether θ or χ is chosen.
Mathematically, a quantity that has the property that its value is determined independently of the method of representation is called a scalar. In other words, conventional definition of the free-energy landscape was a non-scalar quantity. This fact means that the free energy change for the transformation from one stable shape to another one cannot be calculated as an objective physical quantity. The use of conventional definitions did not ensure objectivity in physicochemical discussions and could lead to misinterpretation of data obtained from simulations and experiments in various fields, such as protein folding reactions, catalysis, and drug discovery.
A researcher at AIST has established a method for evaluating free-energy landscape so that it is independent of the representation of the shape.
Free-energy landscape is used in a wide range of fields, such as simulating the expected progress of a reaction of a designed catalyst or predicting drug efficacy and side effects for use in drug development. However, conventional methods derive different free-energy landscapes depending on how the conformational changes of molecules in chemical reactions are represented, and the theoretical basis for quantitative prediction and interpretation has been weak.
In this study, the deformation motion of molecules is expressed by the Langevin equation, which is used to describe Brownian motion. By using the diffusion coefficient that appears in the equation, we succeeded in deriving a free-energy landscape that is independent of the representation of the shape. The results of this research set the theoretical foundation for quantitative discussions of catalytic reactions and protein folding reactions. It is expected that the formulas from this research will be used to provide high quality data on which to base the design of catalysts and pharmaceuticals.