- Japan's Original Genetic Analysis Accelerates Advent of Tailor-Made Medical Care -
The Institute for Biological Resources and Functions (IBRF), of the National Institute of Advanced Industrial Science and Technology (AIST), an independent administrative institution, has developed an innovative technology for genetic quantitative analysis in collaboration with Kankyo Engineering Co.
Until now, the quantitative determination of genes in microbes, animals and plants, the diagnosis of diseases and quality of animals for human consumption, and the analysis of single nucleotide polymorphisms (SNP), indispensable for tailor-made medical care, have demanded resorting to overseas technologies, strictly protected by the intellectual proprietary rights, and for using the technology on business basis, an enormous license fee will be required, urgently demanding the establishment of Japan’s original SNP analysis approach. Under such a circumstance, the joint efforts have led to the development of genetic quantitative and SNP analysis technology of Japanese originality using simple fluorescent quenching probe. The new technology will be applicable not only for R&D purposes but also for practical utilization in various areas such as environment, agriculture, stock raising and medical care. With further advancement, the technology is expected to make a great contribution to the post-genome investigation in Japan.
The AIST Innovation Center for Start-ups (ICS) has positively recognized the feasibility of creating a venture based on these results, and organized a start-up task force under the leadership of a start-up adviser, providing a comprehensive support including urge for additional R&D works toward the commercialization, drafting a business model based on the market research, preparing a business plan and a series of founding processes required for the initiation.
With the overall support from the ICS-AIST, a venture firm named “J-Bio 21” was founded on December 1, 2004, jointly with Kankyo Engineering Co. and Arkray Co., a manufacturer of medical test instruments and reagents for the purpose of spreading the achievements. Subsequently, Dr. Kazunori Nakamura, Deputy Director of the IBRF-AIST took the post of executive officer and the operation was initiated on February 1. J-Bio 21 has already been engaged in a contract project on the synthesis of fluorescent quenching oligo-nucleotides, and succeeded in developing SNP analysis system for diagnosing diseases and quality of cattle of Japanese breed. It will make efforts to expand the business as a proposal type resolution enterprise, specialized in sales of genetic analysis tools and instruments, as well as entrusted analysis and consultant business for genetic analysis based on the accumulated research knowhow, providing technological suggestions and proposals.
In the R&D fields of genetic analysis, the breakthrough development of the polymerase chain reaction (PCR) method of amplifying genes in vitro has been followed by a lot of genetic quantitative analysis approaches utilizing the gene amplification. Most of underlying technologies, however, have been developed in foreign countries, requiring enormous amount of license fee for using them in the industrial application. In view of such situations, it has been claimed that the development of Japan’s original genetic analysis technology is essential for the advancement of biotechnology and the promotion of post-genome studies in Japan.
A five-year project “Development of Utilization Technologies for Composite Biological Systems and Resources” was carried out from fiscal years1997 to 2001 as a part of the “R&D Program of Industrial Science and Technology” program of the New Energy and Industrial Technology Development Organization (NEDO), another independent administrative organization. The project was executed through the collaboration between government and private sectors under the leadership of the National Institute of Bioscience and Human-Technology (NIBH) of former AIST, MITI.
Under this project, the NIBH made a joint research with Kankyo Engineering Co. for developing technologies for analyzing composite biological systems, in particular, an innovative genetic analysis technology. After the completion of NEDO-sponsored project, the IBRF-AIST and Kankyo Engineering Co. have kept working together to develop novel technologies.
Element technologies obtained by the joint effort include the gene detection based on fluorescence quenching effect (QP method) and the single nucleotide polymorphisms (SNP) detection (SNaPpy method) using the fluorescence resonance energy transfer (FRET). The features and implementations of the SNP analysis based on these two technologies will be presented below.
Specific SNP Analysis Technology
The quenching probe (QP) method is to attach a fluorescent marker to a fragment of chemically synthesized gene. When the fragment combines with target nucleic acid, the fluorescence is quenched by the action of guanine (G) in the target, and when the two dissociate, quenched fluorescence restores. In this way, a specific gene can be detected and analyzed. With the restoration of quenched fluorescence of the G-probe, it is possible to detect a slight difference in nucleotide from the thermal dissociation curve (single nucleotide polymorphisms, SNP). The technique requires to attach a single fluorescent probe to a gene fragment, in contrast to the most widely utilized TaqMan method requiring two different probes. The former is characterized by simple design and easiness to use. While the TaqMan method is applicable to the analysis of specific SNP for gene diagnosis, the method is patented to Roche Diagnostics, and a license is needed for using the technique. For this reason, in most of cases of SNP analysis for live stock such as Japanese breed cattle, the analysis had to resort to cumbersome process of cutting products of PCR amplification with specific restriction enzyme and reading gel electrophoresis pattern. The thermal dissociation curve analysis using the Q-probe has been in the process of commercialization as a substitute for the complicated gel electrophoresis. The technique is applicable for the provision of tailor-made medical care based on personal genetic information. [See Commentary 1.]
|
Commentary 1
Specific SNP Analysis Technology (Q Probe Method)
■ Features
1) SNP analysis made available in a single step from the genome DNA through the combination of with gene amplification such as PCR.
2) One single probe is sufficient to analyze one SNP, resulting in sharp reductions in costs for probes.
3) A multiplex of up to two colors can be used, enabling two SNPs to be analyzed using a single tube.
■ Principle
SNP analysis is done by using Q-probe and dissociation curve analysis for amplification product.
Figure. Outline of SNP detection by QP method
Figure. Results of SNP analysis by QP method
|
Large-scale SNP Analysis Technology
The SNaPpy method is a new approach for genotyping SNPs using FRET. Both Q-probe and TaqMan methods, requiring synthesis of gene fragment with fluorescent dye probe for each of SNP session. The SAaPpy method uses fluorescently labeled mononucleotides and DNA-binding fluorogenic molecule as an energy donor for FRET instead of fluorescent probes. This provides a cost merit when a lot of sessions are to be made with a single SNP, but requires enormous cost when doing a limited number of analyses for many SNP specimens (large-scale SNP analysis). For such a purpose, the Invader method has already been developed by Third Wave Technologies Inc.. The SNaPpy method is expected to be a powerful rival for the Invader. The SNaPpy technique will cover every kind of SNP analysis by using four kinds of nucleic acid base (constituents of gene fragment) labeled with fluorescent dye and unlabeled oligonucleotide. In the event when a comprehensive analysis of SNP is needed, the cost and time needed for the synthesis of fluorescent-labeled gene fragments can be extensively reduced. Moreover, as every SNP session is carried out under identical experimental conditions, it is not requested to review condition for each of SNP sessions. It is said that for identifying a causative gene of a certain disease, SNP analysis is needed for tens of thousands sites. The SNaPpy method is best suited for cases where the analysis of a great number of SNP sessions with reasonable cost and simple method is needed. [See Commentary 2.]
Commentary 2
Comprehensive SNP Analysis Technology (SNaPpyTM Method)
--- Innovative Technique for SNP Analysis, Simple and Low Cost ---
■ Features
1) The method uses SYBR Green I and fluorescently-labeled ddNTPs for the FRET reagent instead of fluorescent probes. This can markedly reduce the cost and time for synthesis of fluorescent probes.
2) The method is robust, allowing specific genotyping of most SNPs under the same reaction conditions, as well as simple primer design. The effort required for assay design and optimization is minimal.
3) Genotypes can be determined by end-point fluorescence measurement with a simple optical systems.
4) All reactions can be performed simply in the same vassel by the addition of solutions and incubation.
→ Best suited for large-scale SNP genotyping.
■ Principle
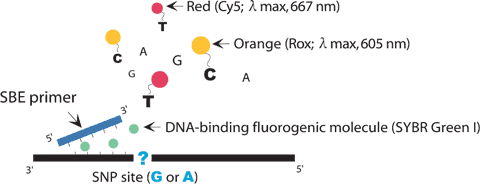
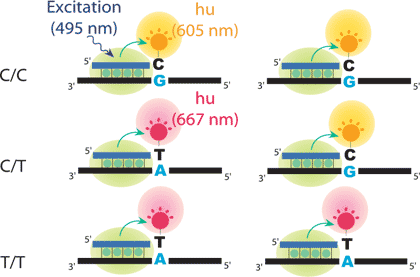
He amplified genomic DNA containing the SNP site is incubated with a SBE primer in the presence of DNA polymerase, SYBR Green I, ddCTP labeled with orange dye, Rox, and ddTTP labeled with red dye, Cy5. The primer binds the complementary site and is extended with a single Rox-ddCTP or Cy5-ddTTP. SYBR Green I binds DNAs and emits green fluorescence on excitation with blue light (495 nm). SBE products containing Rox-ddCTP and Cy5-ddTTP can be excited by the green light emitted by SYBR Green I and emit orange and red fluorescence, respectively, which are measured for SNP discrimination after the SBE reaction.
|