- Restores the native structure of protein and protects it from heat and chemicals -
Naohiro Kameta (Researcher), Organic Nanotube Material Team, the Nanotube Research Center (Director: Sumio Iijima) of the National Institute of Advanced Industrial Science and Technology (AIST; President: Tamotsu Nomakuchi), has developed organic nanotube gels that restore the activity of denatured proteins by folding them into the native three-dimensional structure (refolding). The organic nanotube gels can protect proteins from heat and chemicals.
This technology has been realized by forming organic nanotube gels in which the inner and outer surface structures and diameter of the nanochannel are precisely controlled for the protein of interest. The denatured proteins are encapsulated in the process of the organic nanotube gel formation and are then recovered from the gel by using a change in pH. In this way, only proteins with restored activity can be obtained at high purity. Use of the organic nanotubes as the gel media has the benefit of allowing easy removal of the denaturant by water washing and easy separation and recovery of the protein of interest; these can be achieved without filtration or centrifugal separation. In addition, when proteins are encapsulated in the nanochannel of the organic nanotube, the activity of the proteins is not inactivated by heating or by adding a high concentration of denaturant. Expected applications of this technology include efficient preparation of high-purity proteins and use in nanoreactors and enzyme sensors by combining an enzyme with the organic nanotube.
Details of the results will be published online in ACS Nano, a scientific journal of the American Chemical Society, on May 23, 2012 (JST).
 |
|
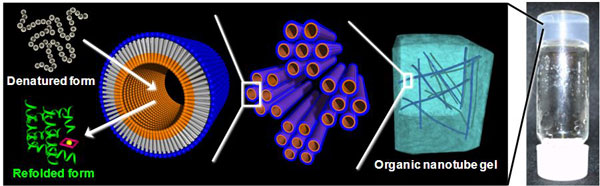
Figure 1 : Refolding of a denatured protein to create a native protein using a nanochannel in the organic nanotube gel |
From a green innovation perspective, enzymes that promote chemical reactions in the body in a specific and highly selective manner with a high yield are attracting substantial attention as energy-efficient, low-environmental-impact catalysts in chemical industrial processes. An enzyme, which is a protein, is folded into a specific three-dimensional structure by the interaction of amino acids (protein components) and exhibits catalytic activity that is based on the structure. Recombinant technology using Escherichia coli is commonly used in the industrial production of proteins. However, an enzyme considerably changes its structure from the native three-dimensional one in the expression of a recombinant protein and aggregates of denatured proteins with no catalytic activity are formed. The restoring efficiency of proteins with a native three-dimensional structure and original catalytic activity from the aggregate is very low. Consequently, various additives to help suppress protein aggregation or to encourage protein refolding into the native three-dimensional structure have been developed. However, the additives have low yield and versatility.
In the body, there is a protein group called molecular chaperones that have nanospaces to encapsulate denatured proteins for isolation and to help with protein refolding. In recent years, porous inorganic materials and polymer nanoparticles that can mimic molecular chaperones have attracted attention. However, an additive is required for desorption of proteins from the porous inorganic materials or polymer nanoparticles and both the additive and incompletely refolded proteins contaminate the aimed protein. Therefore, a complex separation process is required and such a process may cause redenaturation of the protein.
Over more than 10 years, AIST has been working to develop fibrous organic nanomaterials and tubular ones (i.e. organic nanotubes) formed by self-organization in solvents of amphiphilic molecules synthesized from renewable, natural products, such as sugars, amino acids, nucleic acids, and fatty acids. In recent years, AIST has established a process for mass production of organic nanotubes and has been developing various applications of these organic nanotubes.
The outer surface of conventional bilayer-membrane organic nanotubes has the same structure as the inner surface on the nanochannel side. Recently, by using molecular design and molecular arrangement control, AIST has created monolayer-membrane organic nanotubes with an inner-surface structure that differs from that of the outer surface. Providing the nanochannel-side surface with a structure that allows interaction with drugs, protein, DNA, and nanoparticles (called "guests"), the nanotube can encapsulate guests efficiently and selectively. Storage and release of the guests can be regulated by externally controlling the interaction.
The researcher formed organic nanotube gels in which the surface structure and the diameter of the nanochannel were precisely controlled to enable denatured protein molecules to be encapsulated as guests. He also aimed to develop new functions, such as the promotion of protein refolding and the protection of protein activity.
Denatured proteins in which hydrophobic regions are exposed to the surface are prone to aggregation in aqueous solution. A hydrophobic structure was introduced to the nanochannel-side surface to efficiently encapsulate denatured proteins in organic nanotubes and suppress protein aggregation. An amphiphile 1 (Fig. 2a) and its derivative with a hydrophobic benzyl group (shown in green in Fig. 2a) at the end were designed and synthesized, and an organic nanotube gel (L-10-NTG) of the amphiphile 1 and the derivative (molar ratio of 9:1) was formed by self-organization of these two compounds. The gel was formed by controlling pH of the solution with sodium hydroxide at room temperature (Fig. 2b). The organic nanotube consists of a monolayer membrane with molecules of the amphiphile 1 and the derivative arranged in parallel and has an inner diameter of 10 nm (Fig. 2b, c, and d). For comparison an organic nanotube gel H-10-NTG was prepared by self-organization of only amphiphile 1, and an organic nanotube gel H-20-NTG (inner diameter: 20 nm) was prepared by self-organization of only amphiphile 2.
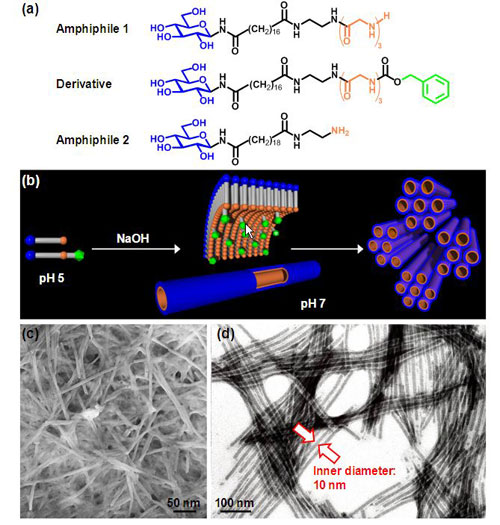 |
Figure 2 : (a) Chemical formulae of the amphiphiles and the derivative;
(b) schematic illustration of the monolayer-membrane organic nanotube;
(c) scanning electron microscope image of the dried organic nanotube gel;
(d) transmission electron microscope image of the organic nanotube. The nanochannels are
visible with phosphotungstate used as a staining reagent. |
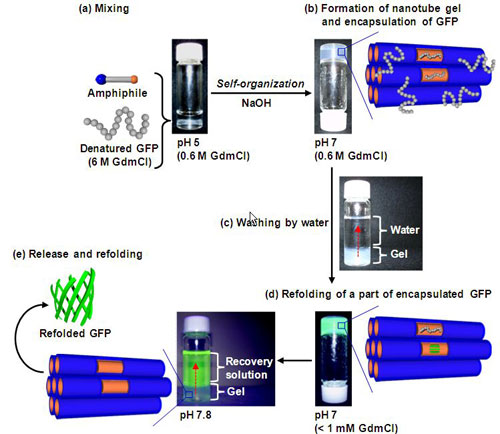
Green fluorescent protein (GFP, molecular weight about 30,000) as a model protein was chemically denatured with guanidinium chloride (GdmCl, concentration 6 M). Denatured GFP was added to the solution during the self-organization of L-10-NTG and H-10-NTG and were encapsulated in the nanochannels when the organic nanotube gels were formed (Fig. 3a, b). The amount of denatured GFP encapsulated in 5 mg of L-10-NTG was 38 µg—about three times more than that encapsulated in H-10-NTG (13 µg). This was probably due to the hydrophobic interaction between the benzyl groups in L-10-NTG and the hydrophobic regions of the denatured GFP.
When the organic nanotube gel encapsulating the denatured GFP was washed with water to dilute the denaturant GdmCl to less than 1 mM (Fig. 3c), some of the encapsulated GFP restored fluorescence activity (Fig. 3d), and protein refolding was promoted in the nanochannels.
 |
|
Figure 3 : Refolding process of GFP denatured by denaturant |
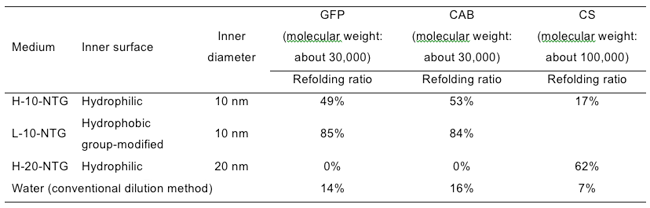
In addition, when a recovery solution (a buffer solution with a pH of 7.8) was added, the glycine amino groups of the amphiphile 1 on the nanochannel surface were uncharged and the electrostatic interaction with the GFP vanished. Consequently, GFP refolded into a native structure in the nanochannels, as well as GFP incompletely refolded in the nanochannels, was recovered in the refolded state from the organic nanotube gel (Fig. 3e). The total refolding ratio of the encapsulated GFP was 49% for H-10-NTG and 85% for L-10-NTG. Introduction of the hydrophobic structure (benzyl groups) into the nanochannels strongly induced refolding, particularly in the release and recovery process (Table 1). The denatured GFP molecules that were not refolded (51% for H-10-NTG and 15% for L-10-NTG) remained encapsulated in the organic nanotube gel; only the refolded GFP was selectively recovered into the recovery solution with adjusted pH without the use of a special additive. The refolding ratio of GFP obtained by the conventional dilution method was 14%, showing that protein refolding was promoted by the organic nanotube gel.
H-10-NTG promoted refolding of carbonic anhydrase (CAB, with a molecular weight of about 30,000–comparable to that of GFP), but it did not efficiently induce refolding of citrate synthase (CS, with a molecular weight of about 100,000–higher than that of GFP) (Table 1). H-20-NTG with an organic nanotube diameter of 20 nm exhibited a high ability to promote refolding of CS, but it did not induce refolding of GFP and CAB at all. Instead, it promoted aggregation of these proteins in the nanochannels. This shows that an organic nanotube gel with an inner diameter appropriate for each protein has a high level of ability to promote protein refolding.
|
Table 1 : Refolding ratios of denatured proteins in the organic nanotube gel systems or the dilution method system |
 |
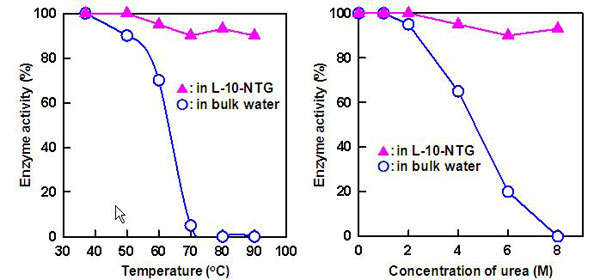
The organic nanotube gels have the ability to promote protein refolding, as well as a protective ability that strongly suppresses the thermal and chemical denaturations of proteins. Whereas free CAB molecules in water lost almost all of their enzyme activity owing to thermal or chemical denaturation by heat or a high concentration of urea (denaturant), the CAB encapsulated in the nanochannels of L-10-NTG were protected from heat or the denaturant and retained 90% of their enzyme activity under the same conditions (Fig. 4).
 |
|
Figure 4 : Comparison of the thermal stability (left) and chemical stability (right) of CAB encapsulated in an organic nanotube gel and free CAB in water |
The researcher intends to create organic nanotube gels in which the surface structure and nanochannel diameter are controlled appropriately for a variety of proteins with different properties. He aims to develop an artificial molecular chaperone system through collaborative research, including provision of samples. He will also apply enzyme-conjugated organic nanotubes to nanoreactors and enzyme sensors.