- The system is readily applicable to the purification of various antibody drugs -
The New Energy and Industrial Technology Development Organization (NEDO), Shimadzu Corporation, the National Institute of Advanced Industrial Science and Technology (AIST), Kyoto Monotech Corporation, and Japan Bioindustry Association (JBA) have jointly developed a protein array analysis system which assesses optimum ligands for purification of various antibodies such as antibody drugs on the basis of their specific characteristics.
Because many proteins can be anchored in a specific direction on a chip, unlike the conventional analysis methods, the new system can simultaneously detect various protein interactions under the same condition as that of chromatography without the need of labeling and can be used for practical screening.
This development is a part of NEDO project entitled "Development of New Functional Antibody Technologies." By using the developed protein array system, we are attempting to establish methods to improve purification processes for the safe and cost-effective antibody drugs.
(1) Because of the silica monoliths, the system can be used to detect protein interactions as well as to perform chromatography.
(2) Use of ultraviolet light absorption enables protein measurement without labeling.
(3) Use of a protein array allows simultaneous detection of multiple protein interactions.
(4) By combining this system with present constituent technologies, we developed a trial set of constituent technologies for antibody purification, which is the final objective of the project entitled "Development of New Functional Antibody Technologies."
This results will be presented at the NEDO booth in Bio Japan 2009, which will be held at Pacifico Yokohama from October 7 to October 9, 2009. Results of other NEDO projects will also be presented at the exhibition. We hope you will come to the exhibits.
For the low-cost production of high-quality antibody drugs, it is necessary to improve the purification processes such as affinity purification. For this purpose, the biggest challenge is to develop an excellent purification column, which can purify a wide variety of antibodies.
NEDO has undertaken a project entitled "Development of New Functional Antibody Technologies" to develop 3 constituent technologies that are necessary to address the above-mentioned issue. The project aims to establish a series of systems by combining the following technologies: ① a technique to design and construct a library that consists of about 103 kinds of affinity ligands, ② a technique to generate high-performance affinity ligands via the following 2 steps: screening the library for affinity ligands that are best suited for a particular antibody that needs to be purified, and then optimizing the affinity ligand, and ③ a technique to develop a support on which the ligands can be fixed.
Constituent technologies mentioned in ① and ③ have already been developed*, and we have developed an array analysis system that achieves the goal mentioned in ②. Technologies for ② primarily involve exhaustive experiments for selecting an optimal ligand from among the 103 kinds of ligands in a library because antibodies are known to have different characteristics. However, this thorough screening requires a very long time. Therefore, we developed a system to rapidly screen high-performance affinity ligands. This system allows the automation of this step, thereby allowing the selection of an optimum ligand from a large ligand library.
Now, a trial set of the constituent technologies has developed. By using the system, we aim to establish technologies for improvement of antibody drugs purification processes, which is the final objective of the NEDO project. The technologies would make antibody drugs cost-effective and safe.
* Development of high-performance affinity supports (beads) for chromatography (press-released by AIST on November 10, 2007) and development of affinity supports (silica monoliths) (press-released by NEDO on January 21, 2009)
In the project entitled "Development of New Functional Antibody Technologies," which has been implemented since FY2006, we developed a cost-effective protein array analysis system that efficiently screens for affinity ligands. In this system, a monolith support is used as a substrate, and various ligands are spotted on the substrate to fabricate an array. The spotted substrate is loaded onto a measurement device to analyze the interactions with proteins, such as antibodies, in each spot. This system consists of (1) a monolith support substrate, (2) a spotter, (3) measurement equipment, and (4) a software program for analysis.
(1) Monolith support substrate
Silica monoliths developed by Kyoto Monotech Corporation are processed into flat plates. Thickness of the standard plate is 0.1 mm so that the influence of light scattering and self-absorption by the silica monolith can be reduced. The silica monoliths have optimized nanometer- and micrometer-sized structures (Fig. 1).
 |
|
Fig1. Monolith support substrate |
(2) Spotter
Shimadzu Corporation developed a spotter to drop samples on the above-mentioned substrate (Fig. 2). This equipment automatically and successively forms 96 spots of ligands by dropping ligand solutions with a volume of several tens of nanoliters. The equipment offers various advantages such as its nozzle can be cleaned before loading different samples to avoid sample mixing. Since the substrate, monolith support, has a structure like micro sponge gourd, the ligand solution vertically penetrates the substrate. With the help of the technology developed by AIST, the C-termini of the affinity ligands can rapidly and specifically crosslink with the monolith support. Another method is also available that allows the crosslinking of the N-termini of the ligands. These technologies allow ligands to be fixed cylindrically in the depth of the spots without being diffused.
 |
|
Fig2. Spotter developed in this project |
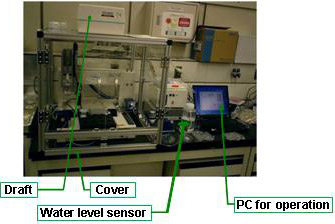
(3) Measurement equipment
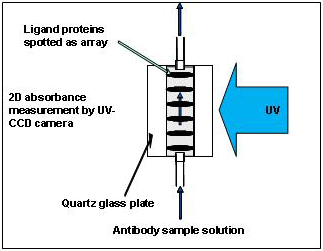
The resultant substrate is placed between 2 quartz glass plates, mounted onto a holder (Fig. 3), and placed into the measurement equipment developed by Shimadzu Corporation (Fig. 4). The substrate is placed between a light source and a photo-detector (CCD camera), and the entire substrate is observed using a CCD camera. The intensity of the light passing through the spotted sites is slightly low because each spot contains the ligand (protein) molecules Note 1, while that of the light passing through other parts of the substrate remains constant (Fig. 5).
When a solution containing proteins, such as antibody molecules, is passed over the flow cell mounted on the equipment, the antibodies interact with the spotted ligands. If the antibodies bind the spotted ligands, the amount of light absorbed increases because the total amount of proteins in the spot increases. In contrast, when the bound antibodies are forcefully detached from the ligands during the subsequent elution process, the amount of light absorbed decreases because of the decrease in the total amount of proteins. If the bound antibodies are washed off completely, the transmitted light is expected to return to the initial state. On the basis of this principle, by fixing different ligands on the 96 spots on a substrate and immersing the ligand array in a solution containing only one kind of antibody, the equipment can simultaneously measure the specificities of all ligands to that antibody, i.e., which ligand-antibody bond is weak and can be easily dissociated and which ligand forms strong bond with the antibody and cannot be dissociated.
(Note 1) If there is no obstacle between the light source and the measurement device (photo-sensor), the photo-sensor receives 100% of the light. However, if there is a substance that scatters or absorbs light, the photo-sensor receives less amount of light. The substrate itself scatters and absorbs light. However, when there are protein molecules on the substrate, they also absorb light, and the photo-sensor eventually receives less light. Because proteins selectively absorb 280 nm of ultraviolet (UV) light, binding and dissociation of proteins can be observed by measuring the amount of absorbed light at this wavelength.
 |
|
Fig3. Monolith support mounted onto a holder |
 |
|
Fig4. Array analysis system developed in this project |
 |
|
Fig5. Mechanism of the measurement (side view of array's cross section) |
(4) Software program for analysis
Images are captured at approximately every 8 seconds to measure the amount of light absorbed. Because it is difficult to analyze the raw images, they are subjected to image processing to measure the amount of light absorbed in each spot. The results of this analysis can be plotted in the form of graphs for better understanding.
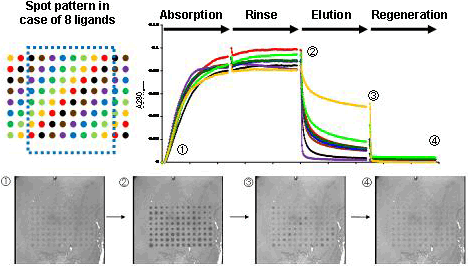
AIST supervised the development and evaluated the system developed. Figure 6 shows a representative evaluation. In this experiment, 8 kinds of ligands were fixed on total 96 spots (upper left panel of Fig. 6); that is, 12 sites of the same color have the same ligand to allow assessment of reproducibility. The substrate was successively immersed in the following solutions: (A) neutral buffer solution containing an antibody (human IgG in this case), (B) neutral buffer solution that did not contain any antibodies, (C) buffer solution at pH 5, and (D) buffer solution at pH 2.5 (plotted on the X axis of the graph in Fig. 6). Variation of 280 nm light intensity at each spot was observed (plotted on the Y axis of the graph in Fig. 6). The following results were obtained: light absorption increased in step (A), was retained in step (B), decreased in step (C), and decreased further and returned to the initial state in step (D). Photographs ①-④ are representative images of spots in each step. After step (D), the support can be reused starting from the step (A). In the above process, step (A) is considered to correspond to adsorption, step (B) to washing, step (C) to elution, and step (D) to recycling. The degree of elution greatly varied among the ligands in step (C). This fact indicates that there is a potential for identifying better ligands in high-throughput analyses.
 |
|
Fig6. An exapmle of resultant data (in case of 8 kinds of ligands) |
(1) Labeling of proteins is not required.
(2) Results can be visually examined through in situ observation.
(3) It is easier to discriminate specific and nonspecific adsorption (in situ observation.)
(4) Evaluation of ligand-antibody interaction properties is possible under chromatography conditions.
(5) High-throughput analyses of elution properties are possible.
(6) Various ligand-antibody interactions can be analyzed.
(7) Arrays can be used (measured) repeatedly.
(8) It has a wide range of prospective applications.
A system based on surface plasmon resonance (SPR) is similar to our system. However, because SPR markedly changes according to the solution conditions such as pH and salt concentration and because in some cases, its signal intensity exceeds the measurement scale, it is difficult to use the SPR-based system under the same measurement conditions as those of chromatography. SPR is, therefore, a useful tool for basic research but is not suitable for measurements intended for practical use. Our system is the automated measurement process based on chromatography that is generally used in laboratories. Therefore, it is suited for measurements intended for practical use. Introduction of this new technology allows users to perform comprehensive research, development, and optimization, including examination of the feasibility of purification (which can increase manufacturing cost), in the earlier phases of development of therapeutic antibodies.