Naohiro Noda (Research Scientist) of the Bio-Measurement Research Group (Leader: Yuji Sekiguchi), the Institute for Biological Resources and Functions (Director: Masahiro Iwakura) of the National Institute of Advanced Industrial Science and Technology (AIST; President: Hiroyuki Yoshikawa) (hereinafter referred to as AIST), in collaboration with Satoshi Tsuneda (Professor) of the Department of Science and Engineering of Waseda University (President: Katsuhiko Shirai), Hidenori Tani of the Graduate School of Waseda University, and Shinya Kurata (Technical Director) of J-Bio21 Corporation (President: Toshifumi Kodama) have developed a new technique — the alternately binding quenching probe competitive assay combined with loop-mediated isothermal amplification (ABC-LAMP) method — for simple, accurate, and inexpensive DNA analysis.
The techniques for quantifying DNA are important for society because they can be used for the analysis of DNA expression in order to diagnose human diseases, for the detection and quantification of viruses such as the severe acute respiratory syndrome (SARS) and avian influenza viruses that cause life-threatening diseases, and for the analysis of mixtures containing genetically engineered foods such as genetically modified soybean.
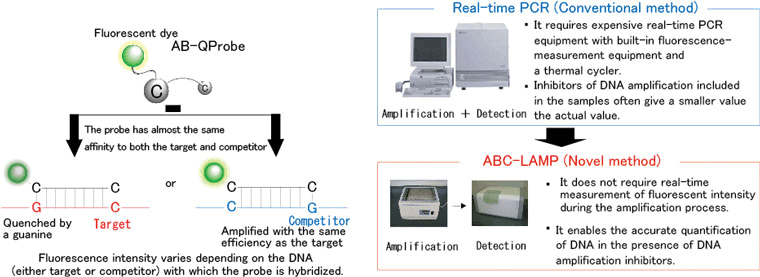
This newly developed technique realizes accurate quantitative analysis of DNA by combining a probe (AB-QProbe) labeled with a dye whose fluorescence is quenched by interaction with guanine (one of the 4 bases that comprise DNA) and an internal standard comprising a known concentration of DNA (competitor). This allows easy and inexpensive DNA analysis because the quantity of target DNA can be determined merely by measuring the fluorescence intensity of the sample before and after DNA amplification (Figure 1).
Compared to the conventional technique for DNA analysis, our novel method has the following advantages: it does not require real-time measurement of fluorescence intensity during amplification, gel electrophoresis, and expensive equipment for real-time polymerase chain reaction (PCR); and it is barely affected by substances that inhibit DNA amplification.
The details of this technique will be released in the online version of Analytical Chemistry published by the American Chemical Society.
 |
|
Figure 1 Schematic presentation of ABC-LAMP and its comparison with the conventional real-time PCR method |
The technique for the quantitative analysis of DNA is important to society because it is used for the analysis of DNA expression in order to diagnose human diseases, for the detection and quantification of viruses such the SARS and avian influenza viruses that cause life-threatening diseases, and for the quantitative analysis of mixtures containing genetically engineered foods such as genetically modified soybean.
Real-time PCR is currently used to quantitatively analyze DNA in microorganisms, plants and animals. In this method, DNA is amplified by PCR, and the amount of amplified DNA is measured during each PCR cycle. Because real-time PCR requires measurement of the fluorescence intensity of the amplification product during each cycle, it entails several problems including the following: (1) it requires expensive real-time PCR equipment with built-in fluorescence measurement equipment and a thermal cycler, (2) its yield of a high throughput (high-speed mass processing) is limited, and (3) it underestimates the quantity of DNA in the presence of any substance that inhibits DNA amplification.
The development of a simple, inexpensive, accurate, high-throughput method that can overcome these problems is greatly desired, not only in the sphere of life sciences but also in various industrial fields including environmental, agricultural, and food sciences.
AIST has been promoting the development of such a method for the quantitative analysis of DNA. As a part of this endeavor, AIST has been collaborating with Waseda University and J-Bio21 Corporation for the development of a new technique involving the fluorescence-quenching phenomenon. Through this collaboration, we successfully developed a new method using a fluorescent dye whose fluorescence is quenched by specific interaction with the guanine bases in DNA; we also developed a competitor (internal standard) containing unique sequences.
This research was financially supported by Industrial Technology Research Grant Program in 2006 from New Energy and Industrial Technology Development Organization (NEDO) of Japan.
The new method of analysis is based on the combined use of the developed probe (AB-QProbe) with a fluorescent dye, which is quenched through its specific interaction with the guanine bases, and a competitor DNA (internal standard). Target DNA can be quantified merely by measuring the fluorescence intensity before and after DNA amplification (Figure 1). The characteristic features of the method are as follows: it does not require real-time measurement of the fluorescence intensity during amplification, gel electrophoresis, or expensive PCR equipment; and it allows accurate analysis because it is barely affected by substances that inhibit DNA amplification.
We incorporated the probe and competitor into the LAMP procedure, an already established DNA amplification method. Thus, we developed a simple, accurate, and inexpensive method, namely, “Alternately Binding Probe Competitive-LAMP” (ABC-LAMP) for the quantitative analysis of DNA.
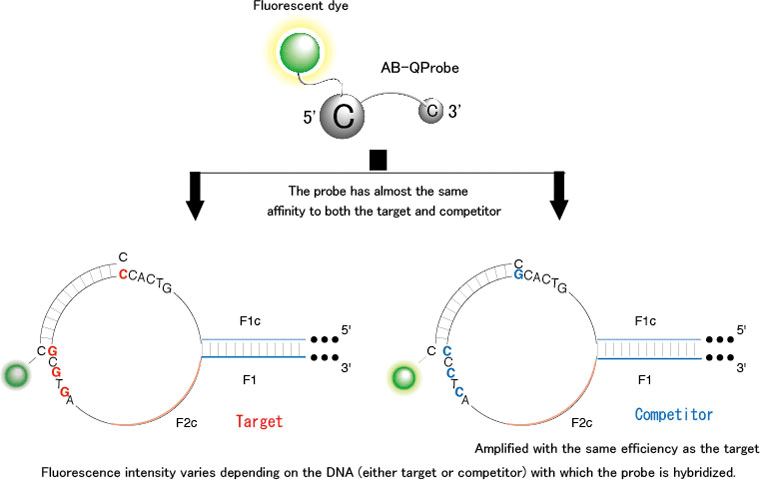
This method used the AB-QProbe along with the target DNA and competitor. Each molecule in the AB-QProbe carries a fluorescently labeled cytosine (C) base at its end; the fluorescence of this base is quenched when it encounters a guanine (G) base on the target DNA. The competitor is amplified with the same efficiency as the target DNA, and it has been modified to possess a C base instead of a G base at the end of the complementary part to which the probe binds This prevents the quenching of the fluorescence of the probe during hybridization with the competitor.
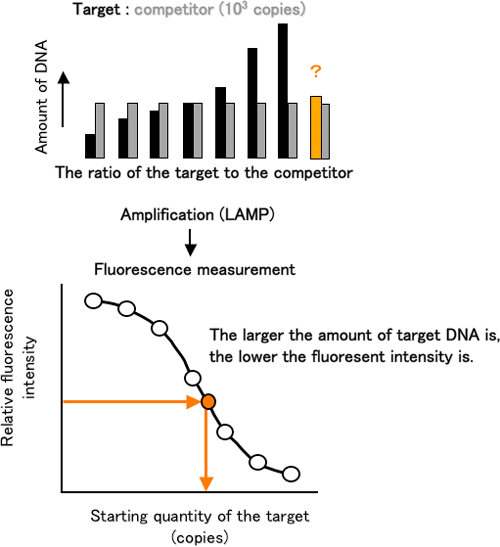
The AB-QProbe competitively hybridizes with the amplification products derived from both the target DNA and competitor with the same affinity; however the fluorescence quenching efficiency will vary depending on the DNA (either target or competitor) with which the probe is hybridized (Figure 2). From the difference in the quenching efficiency, we can quantitatively analyze the target DNA by measuring the fluorescence intensity before and after the LAMP (Figure 3).
 |
|
Figure 2 Schematic presentation of the quenching pattern of the AB-QProbe |
 |
|
Figure 3 Schematic presentation of the quantification of a specific gene by using ABC-LAMP |
This method can be applied not only to LAMP but also to other amplification systems such as PCR. Moreover, the nucleic acid sequence-based amplification (NASBA) method, wherein specific RNAs are amplified, is currently under development. Since our new method measures the fluorescence intensity before and after the amplification reaction, it will be easy to combine it with these amplification methods. We are eagerly awaiting further developments in these technologies.
Using our new method, we determined the quantity of a region of DNA that encoded an ammonia-oxidizing enzyme from ammonia-oxidizing bacteria, and we observed that our method has the same precision and reproducibility as the conventional technique. In the presence of increased concentrations of the DNA amplification inhibitors, humic acid and urea, the conventional method yielded values lower than the true value; however, our method yielded accurate measurements irrespective of these inhibitors.
We will attempt to apply this technique to the analysis of DNA expression for the diagnosis of human diseases and for the qualitative detection of influenza and SARS viruses. J-Bio21 Corporation, an AIST-accredited venture company, is developing inexpensive fluorescence-measuring equipment optimized for this method.