Update(MM/DD/YYYY):04/15/2020
Gene Sequence Design Technology for Increasing Protein Production of Microorganism
– Using information technology to accelerate bio-manufacturing –
Researchers: SAITO Yutaka, Senior Researcher, Computational Omics Research Team, Artificial Intelligence Research Center, also Biotechnology Research Institute for Drug Discovery, also AIST-Waseda University Computational Bio Big-Data Open Innovation Laboratory (CBBD-OIL), KAMEDA Tomoshi, Senior Researcher, Computational Omics Research Team, also Biotechnology Research Institute for Drug Discovery, KITAGAWA Wataru, Senior Researcher, Applied Molecular Microbiology Research Group, Bioproduction Research Institute, also CBBD-OIL, and TAMURA Tomohiro, Deputy Director General, Department of Life Science and Biotechnology, also CBBD-OIL
Summary
The researchers have developed a gene sequence design method based on information technology. They applied the method to protein production using actinomycetes, and demonstrated a high probability of increased production.
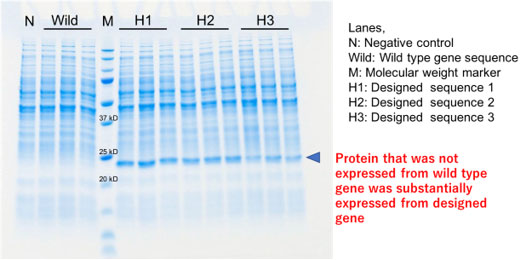
Example of enhanced expression level by sequence design (Protein electrophoresis analysis)
Background
In material production using microorganisms, heterologous genes are artificially introduced into the target microorganism to have the microorganism produce proteins it does not naturally possess. Technology for properly designing the DNA sequence of the genes to be introduced (codon optimization) is important for increasing production amounts. However, codon optimization methods have not been established for the bioindustry-relevant microorganisms, and development is demanded.
New results
The developed method examines only the beginning of the gene sequence instead of the overall sequence. This makes it possible to greatly reduce the number of gene sequences to be examined, which enables computations. The researchers designed the optimal gene sequences for twelve genes and introduced them to an Actinomycete, Rhodococcus erythropolis. Nine of these genes were confirmed to increase production compared to the wild-type sequences. The developed method has several advantages including simple and inexpensive preparation of the gene, and it is potentially applicable to various microorganisms.
Future plans
The researchers will apply this method to the protein production of various microorganisms to demonstrate its effectiveness and try to increase the efficiency of material production.